News Archive
Complementing conventional antibiotics
May 2018. Antimicrobial resistance (AMR) is a major medical problem worldwide, impacting both human health and economic well-being. A new strategy for fighting bacteria has now been reported in the latest online issue of Nature by a research group headed by Ivan Dikic at Goethe University Frankfurt. The scientists revealed the molecular action mechanism of a Legionella toxin and developed a first inhibitor.
As resistance continues to spread, common infections such as pneumonia and salmonellosis are becoming increasingly harder to treat. Two factors drive the AMR crisis: human negligence in the use of antibiotics and a lack of truly novel antibiotics for more than 30 years. According to a recent report by the World Bank, by 2050, AMR may reduce the global gross domestic product by 1.1% to 3.8%, depending on which scenario plays out.
Scientific efforts are underway to achieve better control of microbial infections. One promising approach is to limit damage to host cells and tissues in the course of a bacterial infection by blocking the microbial processes that cause such damage. The laboratory of Ivan Dikic, Director of the Institute of Biochemistry II at Goethe University, has been working in this field for the past decade. As Dikic explains: “We believe we can find new treatments that complement conventional antibiotics by targeting specific groups of bacterial effectors with rationally designed drugs. In this way pathogenic damage can be decreased, which helps patients better tolerate bacterial infection. This is a relatively new field that is attracting more and more attention in the community.”
 To prove that this strategy is a viable option for tackling bacteria, Dikic’s team studies Legionella, which are known to cause pneumonia and are especially dangerous for immunocompromised patients. Recently, the Frankfurt scientists were involved in identifying a novel enzymatic mechanism that Legionella bacteria use to seize control over their host cells. Sagar Bhogaraju, who works at Goethe University’s Buchmann Institute for Molecular Life Sciences as part of the Dikic team, reports: “We showed that Legionella enzyme SdeA acts as a toxic bacterial effector. It promotes the spreading of bacteria by targeting the ubiquitin system, one of the cell’s powerful protection mechanisms against stress.”
To prove that this strategy is a viable option for tackling bacteria, Dikic’s team studies Legionella, which are known to cause pneumonia and are especially dangerous for immunocompromised patients. Recently, the Frankfurt scientists were involved in identifying a novel enzymatic mechanism that Legionella bacteria use to seize control over their host cells. Sagar Bhogaraju, who works at Goethe University’s Buchmann Institute for Molecular Life Sciences as part of the Dikic team, reports: “We showed that Legionella enzyme SdeA acts as a toxic bacterial effector. It promotes the spreading of bacteria by targeting the ubiquitin system, one of the cell’s powerful protection mechanisms against stress.”
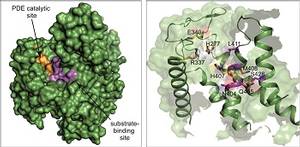
Ivan Dikic’s group has now reported a further breakthrough in the journal Nature: they succeeded in solving the atomic structure of SdeA. “The enzyme is truly unique and catalyses a reaction in a two-step mechanism”, comments Sissy Kalayil, who is one of the lead Frankfurt scientists on the project. “Our results are very exciting as they reveal atomic details of this mechanism, and make the rationale design of inhibitors possible.” In their publication, the researchers also reveal how this bacterial effector probably chooses its victim proteins within the host cell, exerting its effect by attaching ubiquitin to them. They also developed a first inhibitor blocking this reaction in vitro. “Our basic discovery has allowed us to prove that these enzymes are druggable,” Dikic comments. “But it is early days. There is a long road ahead of us before we will be able to use this novel mechanism therapeutically. And we will surely not stop here.” Most likely, Legionella is not the only bacterium using this mechanism.
Ivan Dikic’s group is located at the Institute of Biochemistry II (Medical Faculty) and the Buchmann Institute for Molecular Life Sciences at Goethe University Frankfurt. The group investigates the role of ubiquitin in human diseases including cancer, amyotrophic lateral sclerosis and infectious diseases.
Publication:
Kalayil S*, Bhogaraju S*, Bonn F, Shin D, Liu Y, Gan N, Basquin J, Grumati P, Luo Z-Q, Dikic I. Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination. Nature, advanced online publication 23 May 2018, DOI 10.1038/s41586-018-0145-8 (* shared first authorship) Link
In the same issue of Nature, there will be two articles published by the groups of Yue Feng (China) and Yuxin Mao (USA) which contribute additional details to the molecular mechanism of this unique enzyme (DOI 10.1038/s41586-018-0146-7 und 10.1038/s41586-018-0147-6
Contact:
Kerstin Koch, Institute of Biochemistry II, University Hospital Frankfurt, Frankfurt am Main, Germany, Tel. +49 69 6301 84250, k.koch@em.uni-frankfurt.de
Figure caption: A detailed view of the 3D structure of the enzymatic active part of SdeA toxin (green). On the left, the essential catalytic surface is depicted in orange and the area for binding target proteins in purple. On the right, the amino acid residues involved in the reaction are highlighted. The detailed molecular picture now enables the design of suitable inhibitors. Source: Nature/Kalayil et al, Mai 2018
Cluster of Excellence Macromolecular Complexes, Frankfurt am Main, Germany

