News Archive
New insights into the dynamics of riboswitch activation
December 2018. Riboswitches are important regulatory segments of messenger RNA that bind particular small molecules (ligands), resulting in a change in production of the protein encoded by the mRNA. A recently published study led by Josef Wachtveitl from Goethe University Frankfurt and Snorri Sigurdsson from the University of Iceland reveals important new details of how riboswitches bind ligands.
Each riboswitch has an ‘aptamer domain’ and an ‘expression platform’. The small molecule ligand binds to the aptamer domain resulting in a change of the structure of the expression platform, which regulates gene expression. The aptamer is central for the overall function and binds the ligand with an extraordinary high affinity and specificity. It is therefore of great interest to understand such binding motifs and to obtain a molecular picture of the mechanisms and dynamics of the binding process.
RNA aptamers for a wide range of ligands can be produced artificially using in vitro selection. An example is the N1 aptamer, which is one of the smallest known aptamers that binds with high affinity to the antibiotic neomycin. This aptamer has an internal loop and a terminal loop which are connected by a short helical stem region.
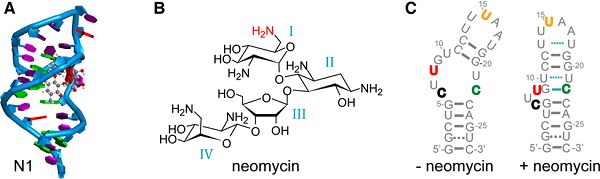
Figure legend: (A) NMR structure of the N1 neomycin RNA aptamer with bound ligand (here ribostamycin).
(B) Structure of the antibiotic neomycin B. The NH2-group marked in red contributes to the H-bonding pattern of the neomycin aptamer.
(C) Predicted structures of the ligand unbound (left) and bound (right) state of the neomycin RNA aptamer. Bold-colored letters for the RNA bases mark the different Çmf labeling positions. (Nucleic Acids Res. 2018. doi:10.1093/nar/gky1110)
Structural information is indispensable for understanding RNA–ligand recognition. NMR and electron paramagnetic resonance (EPR) spectroscopic studies have shown that the binding pocket of N1 is preformed and that the antibiotic is bound to the aptamer via a conformational selection mechanism. However, the picture is not complete without information on the dynamics of the aptamer.
To investigate this further, the scientists used fluorescence spectroscopy. This method provides information on quantum yield, lifetime or anisotropy, giving deeper insights into micro-environment changes upon ligand binding at a defined position within the RNA. The method requires fluorescent reporter labels (fluorophores) since neither RNA nor most ligands are fluorescent.
Fluorescent labels have to meet several stringent requirements. In particular, it should be possible to incorporate them site-specifically. The team selected for their study a new reporter label called Çmf, which is based on the cytidine analogue Ç. Cytidine is one of the main building blocks of of RNA and DNA. A similar analogue, Çm, which contains a 2′-methoxy group, was recently established as a label for EPR studies of RNA. Ç and Çm can be modified with a reducing agent to yield the fluorescent nucleosides Çf and Çmf, respectively. The new study is the first time Çmf has been used as a fluorescent label in RNA research. The team conducted a detailed characterization of Çmf for both steady-state and time-resolved fluorescence measurements.
For the ligand binding study the N1 aptamer was labelled at four different positions. With steady-state fluorescence methods, it was possible to confirm the conformational selection mechanism. It was possible to observe the dynamics of the ligand binding process with the help of fluorescence-monitored stopped-flow measurements. It became clear that the ligand binding to the aptamer is in fact a two-step process. The first step is an unspecific ligand binding near or in the binding pocket and in a second step the ligand is bound specifically with the help of H-bonds and electrostatic interactions.
Fluorescence lifetime measurements allowed the scientists to distinguish between labeled double- and single-strands and also between flanking bases of the fluorophore. The Çmf fluorophore is sensitive to its micro-environment such as base pairing, stacking and solvent accessibility. Thus, specific Çmf-labelled RNA samples are perfectly suited to duplexation and ligand binding studies. Time-resolved fluorescence measurements of Çmf-labelled samples allow structural dynamics studies. The results thus serve as a benchmark for analogous experiments on functional RNAs.
Contact:
Josef Wachtveitl, Institute of Physical and Theoretical Chemistry, Riedberg Campus, Goethe University, Frankfurt/Main, Germany, wveitl@theochem.uni-frankfurt.de
Publication:
Henrik Gustmann, Anna-Lena Segler, Dnyaneshwar Gophane, Andreas Reuss, Christian Grünewald, Markus Braun, Julia Weigand, Snorri Th Sigurdsson* and Josef Wachtveitl* (2018) Structure guided fluorescence labeling reveals a two-step binding mechanism of neomycin to its RNA aptamer. Nucleic Acids Research: gky1110, published online 20 November 2018, dx.doi.org/10.1093/nar/gky1110
Cluster of Excellence Macromolecular Complexes, Frankfurt am Main, Germany

