News Archive
Structure and autoregulation of a flippase that drives active transport of phospholipids
July 2019. An international team of scientists from the Max Planck Institute of Biophysics in Frankfurt, the Université Paris-Sud in France and Aarhus University in Denmark have solved the structure and regulation of an important cellular flippase protein complex.
Cells and organelles are enclosed by lipid-bilayers and membrane proteins. In eukaryotic membranes that are involved in the secretory and endocytic pathways, the lipid distributions between the two bilayer leaflets are asymmetric. These lipid gradients potentiate key biological processes such as membrane dynamics, endocytosis, exocytosis and signalling.
This lipid asymmetry needs to be constantly regulated. Members of two distinct superfamilies of membrane proteins drive the ATP-dependent unidirectional translocation of lipids against concentration gradients: ATP-binding cassette transporters typically drive the inward-to-outward (flop) translocation of lipids between bilayer leaflets, whereas P4-ATPases drive the outward-to-inward (flip) process.
Although previous work shed light on the structure and mechanism of lipid floppases, P4-ATPases had been studied only through bioinformatics and functional assays. Models had identified potential peripheral or centrally located lipid-recognition sites and pathways, but the molecular architecture of P4-ATPases and the mechanism through which they recognize and transport lipids remained unknown.
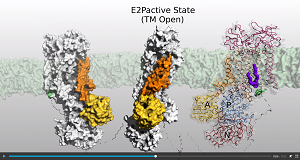 To gain structural and mechanistic insights, the scientists embarked on cryo-electron microscopy studies of a P4-ATPase from yeast called Drs2p–Cdc50p. They were successful in obtaining high resolution 3D structures that capture the progressive steps from a fully autoinhibited flippase, to an intermediate activated state as well as an outward-open and activated conformation. Their results were published in the journal Nature.
To gain structural and mechanistic insights, the scientists embarked on cryo-electron microscopy studies of a P4-ATPase from yeast called Drs2p–Cdc50p. They were successful in obtaining high resolution 3D structures that capture the progressive steps from a fully autoinhibited flippase, to an intermediate activated state as well as an outward-open and activated conformation. Their results were published in the journal Nature.
The investigated flippase is specific to phosphatidylserine and phosphatidylethanolamine lipids and is autoinhibited by the C-terminal tail of Drs2p. It is activated by the lipid phosphatidylinositol-4-phosphate. The new study highlights specific features of P4-ATPases and reveals sites of autoinhibition and PI4P-dependent activation. The scientists also observed a putative lipid translocation pathway in this flippase that involves a conserved PISL motif in transmembrane segment 4 and polar residues of transmembrane segments 2 and 5, in particular Lys1018, in the centre of the lipid bilayer. Further information ...
Contact:
Arne Möller, Max Planck Institute of Biophysics, Frankfurt/Main, Germany, arne.moeller@biophys.mpg.de
Publication:
Milena Timcenko, Joseph A. Lyons, Dovile Januliene, Jakob J. Ulstrup, Thibaud Dieudonné, Cédric Montigny, Miriam-Rose Ash, Jesper Lykkegaard Karlsen, Thomas Boesen, Werner Kühlbrandt, Guillaume Lenoir*, Arne Moeller* & Poul Nissen* (2019) Structure and autoregulation of a P4-ATPase lipid flippase. Nature: published online 26 June 2019. http://dx.doi.org/10.1038/s41586-019-1344-7

