News Archive
Life times of metastable states guide regulatory signaling in transcriptional riboswitches
March 2018. Riboswitches represent an important class of regulatory RNA elements. Metabolite binding to transcriptional riboswitches can lead to either premature termination of transcription (“OFF switch”) or continuation of transcription (“ON switch”). It has been shown that many transcriptional riboswitches regulate under kinetic control, where both ligand binding and the formation of secondary structures that either induce or escape termination (terminator/antiterminator) are cotranscriptional events.
Transcriptional riboswitches modulate downstream gene expression by a tight coupling of ligand-dependent RNA folding kinetics with the rate of transcription. RNA folding pathways leading to functional ON and OFF regulation involve the formation of metastable states within well-defined sequence intervals during transcription. The kinetic requirements for the formation and preservation of these metastable states in the context of transcription remain unresolved.
Scientists at Goethe University Frankfurt and the University of Wisconsin–Madison now report a new study in which they managed to reversibly trap the regulatory relevant metastable intermediate of a riboswitch using a photocaging-ligation approach and to monitor folding to its native state by real-time NMR in both presence and absence of ligand. The riboswitch used was the 2′dG-sensing riboswitch from the bacterium Mesoplasma florum. The scientists also determined transcription rates for two different bacterial RNA polymerases.
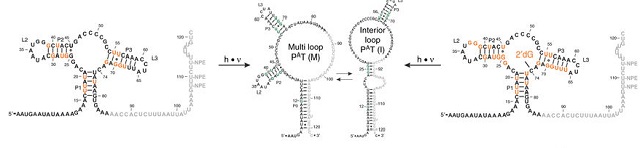
Their results reveal that the riboswitch functions only at transcription rates typical for bacterial polymerases (10–50 nucleotides per second) and that gene expression is modulated by 40–50% only, while subtle differences in folding rates guide population ratios within the structural ensemble to a specific regulatory outcome.
General fluctuations in transcription rates resulting from changes in cellular conditions may be self-regulated by the negative feedback loop of the riboswitch itself. The kinetic and simulation framework developed by this study provides an important contribution to the conceptual basis to understand transcriptional riboswitches. More...
Contacts:
Harald Schwalbe and Alexander Heckel, Institute of Organic Chemistry and Chemical Biology, Goethe University Frankfurt, Germany, schwalbe@nmr.uni-frankfurt.de, heckel@em.uni-frankfurt.de
Publication:
Helmling C, Klötzner DP, Sochor F, Mooney RA, Wacker A, Landick R, Fürtig B, Heckel A*, Schwalbe H* (2018) Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nature Communications 9: 944. http://dx.doi.org/10.1038/s41467-018-03375-w

