News Archive
High quality photoswitchable regulatory RNA tool developed
December 2018. Scientists from the universities of Frankfurt and Darmstadt have jointly developed an exciting new tool to investigate and regulate complex molecular biological networks. The new tool is a component of a riboswitch (a type of regulatory RNA), and consists of two parts: an RNA sequence called an aptamer and a small molecule (ligand) that can bind to this aptamer. What makes this aptamer-ligand combination special is that the binding of the ligand to the aptamer can be switched on and off by light, thus helping scientists to obtain precise control over the process they are interested in.
In recent years, biologists and biochemists have become increasingly interested in light-controlled nucleotide tools. Light can be controlled spatially and temporally. In biological systems, light can be used as an external independent trigger because most cells do not respond to light (except photoreceptors). Many cells are at least partially translucent and they are not damaged by light if it is used at the right wavelength. Some organisms such as nematode worms and young zebra fish are translucent and both are important model species for research.
RNA riboswitches are an excellent example of RNA-based regulation. They are highly structured naturally occurring RNA elements that form a binding pocket with their aptamer domain, which can recognize metabolites with high affinity and specificity. The ligand binding to this pocket affects a change in a second domain of the riboswitch, called the the expression platform, which then causes a change in gene expression. A number of synthetic riboswitches have been developed that are modelled on natural riboswitches but use as binding domains novel aptamers. These aptamers are selected in the laboratory with a method called SELEX. With this method aptamers can be selected against almost any ligand of choice. Synthetic riboswitches are part of the toolbox of many synthetic biology applications.
The new tool reported here is a novel kind of RNA aptamer, whose binding depends on a light-induced change of conformation of its purpose-designed small molecule ligand (called ‘azoCm’). The scientists chose an azobenzene as the basis of azoCm because of its reliable photoswitchability and modified it with chloramphenicol for a better interaction with RNA. The synthesis of azoCm was followed by extensive biophysical analyses regarding its stability and photoswitchability. RNA aptamers were identified after several cycles of selection in the laboratory and then studied regarding their binding specificity and affinity toward the ligand. The team succeeded in developing an RNA aptamer that selectively binds to only the trans photoisomer of azoCm with a dissociation constant of 545 nM. When the ligend is exposed to light of a wavelength of 365 nm, the aptamer cannot bind the ligand anymore because the ligand changed its structure (→ figure). The binding can be "switched on" again by exposing the ligand to light of a wavelength of 420 nm.
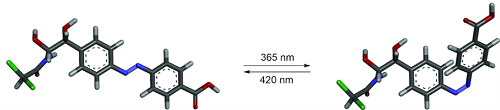
azoCm is non-toxic, stable in biological medium and can be photoswitched multiple times without significant degradation and with high photoswitching amplitude. High-quantum yields, low-thermal rates of conversion and spectrally addressable photoisomers make this ligand ideal for applications in a biological context.
Contacts:
Alexander Heckel, Institute of Organic Chemistry and Chemical Biology, heckel@em.uni-frankfurt.de
Josef Wachtveitl, Institute of Physical and Theoretical Chemistry, wveitl@theochem.uni-frankfurt.de
Riedberg Campus, Goethe University, Frankfurt/Main, Germany
Publication:
Thea Lotz, Thomas Halbritter, Christoph Kaiser, Martin Rudolph, Leon Kraus, Florian Groher, Sabrina Steinwand, Josef Wachtveitl*, Alexander Heckel* & Beatrix Suess* (2018) A light-responsive RNA aptamer for an azobenzene derivative. Nucleic Acids Research: gky1225, published online 5 December 2018. http://dx.doi.org/10.1093/nar/gky1225
Cluster of Excellence Macromolecular Complexes, Frankfurt am Main, Germany

