News Archive
High resolution structure of a bacterial terminal oxidase provides clues for antimicrobial drug development
October 2019. Cytochrome bd–type quinol oxidases are enzymes that catalyze the reduction of molecular oxygen to water in the respiratory chain of many human-pathogenic bacteria. They are structurally unrelated to human mitochondrial cytochrome c oxidases and are therefore a prime target for the development of antimicrobial drugs.
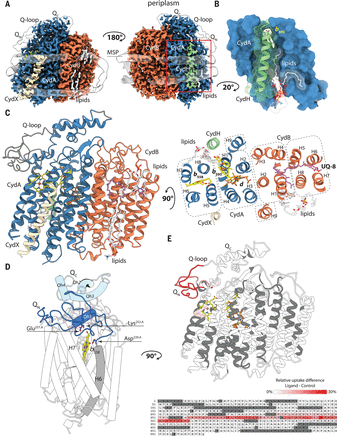 Scientists at the Max Planck Institute of Biophysics in Frankfurt have succeeded in determining the structure of the Escherichia coli cytochrome bd-I oxidase by single-particle cryo–electron microscopy to a resolution of 2.7 angstroms. Their structure, published on 4th of October 2019 in the journal Science, contains a previously unknown accessory subunit CydH, the L-subfamily–specific Q-loop domain, a structural ubiquinone-8 cofactor, an active-site density interpreted as dioxygen, distinct water-filled proton channels, and an oxygen-conducting pathway. Comparison with another cytochrome bd oxidase revealed structural divergence in the family, including rearrangement of high-spin hemes and conformational adaption of a transmembrane helix to generate a distinct oxygen-binding site.
Scientists at the Max Planck Institute of Biophysics in Frankfurt have succeeded in determining the structure of the Escherichia coli cytochrome bd-I oxidase by single-particle cryo–electron microscopy to a resolution of 2.7 angstroms. Their structure, published on 4th of October 2019 in the journal Science, contains a previously unknown accessory subunit CydH, the L-subfamily–specific Q-loop domain, a structural ubiquinone-8 cofactor, an active-site density interpreted as dioxygen, distinct water-filled proton channels, and an oxygen-conducting pathway. Comparison with another cytochrome bd oxidase revealed structural divergence in the family, including rearrangement of high-spin hemes and conformational adaption of a transmembrane helix to generate a distinct oxygen-binding site.
In case of the E. coli bd-I oxidase, it may be possible to design inhibitors that compete with UQ-8 for the CydB binding site and interfere with the assembly of a functional CydAB core dimer. An alternative target is the membrane-embedded entry site of the oxygen-conducting O-channel that would be targeted by hydrophobic compounds to block the channel and abolish oxygen uptake. Further structural studies of bd oxidases are needed to explore whether the observed structural differences between E. coli and Geobacillus thermodenitrificans are specific for their respective bacterial clades or result from a distinct adaptation to their ecological niche and physiological roles.
Contacts:
Werner Kühlbrandt (werner.kuehlbrandt@biophys.mpg.de) and Hartmut Michel (michel@mpibp-frankfurt.mpg.de) Max Planck Institute of Biophysics, Frankfurt/Main, Germany
Publication:
Safarian S, Hahn A, Mills DJ, Radloff M, Eisinger ML, Nikolaev A, Meier-Credo J, Melin F, Miyoshi H, Gennis RB, Sakamoto J, Langer JD, Hellwig P, Kühlbrandt W*, Michel H* (2019) Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science 366: 100-104. http://dx.doi.org/10.1126/science.aay0967

