News Archive
A chemical toolbox for the study of epigenetic signaling
May 2019. Genetic and epigenetic variation, as well as environmental and lifestyle factors, work in concert to influence human health and disease. In recent years, the essential role of epigenetic modifications in regulating gene expression and cellular differentiation has emerged. Apart from changes in DNA methylation, covalent post-translational modifications of histones and other nuclear proteins define a complex language, the epigenetic code, which regulates chromatin structure and dynamics.
Lysine acetylation is a major epigenetic post-translational modification occurring on histone proteins. This type of modification has generally been associated with activation of transcription through opening of chromatin structure, although some recent studies have found some lysine acetylation marks to be responsible for the compaction of chromatin, protein stability, and the regulation of protein-protein interactions.
Disruption of histone acetylation patterns has been linked to the development of disease. This may occur through mutations that deregulate enzymes responsible for adding or removing these histone acetyl marks, as well as the protein interaction modules that recognize and interpret this important post-translational modification.
The complex pattern of acetylation marks is interpreted by reader domains of the bromodomain family of proteins. Bromodomain-containing proteins have been conserved during evolution. There are 61 bromodomains expressed in the human proteome, present in 46 diverse proteins. However, the roles of bromodomains in regulating cellular states and their potential as targets for the development of targeted treatment strategies is poorly understood.
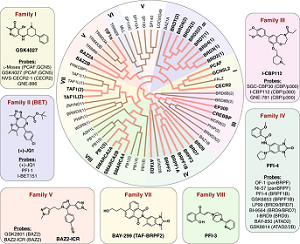 Scientists from Goethe University in collaboration with scientists from Toronto, Ottawa and Oxford have developed a set of 25 chemical probes, selective small molecule inhibitors, covering 29 human bromodomain targets. The team comprehensively evaluated the selectivity of this probe-set and demonstrated the utility of the set identifying roles of bromodomains in cellular processes and potential translational applications. They discovered, for example, crosstalk between histone acetylation and the glycolytic pathway resulting in a vulnerability of breast cancer cell lines to inhibition by certain bromodomains.
Scientists from Goethe University in collaboration with scientists from Toronto, Ottawa and Oxford have developed a set of 25 chemical probes, selective small molecule inhibitors, covering 29 human bromodomain targets. The team comprehensively evaluated the selectivity of this probe-set and demonstrated the utility of the set identifying roles of bromodomains in cellular processes and potential translational applications. They discovered, for example, crosstalk between histone acetylation and the glycolytic pathway resulting in a vulnerability of breast cancer cell lines to inhibition by certain bromodomains.
This chemical probe-set will aid the elucidation of further roles of bromodomain proteins in normal and disease states. It is also an excellent toolset for the validation of bromodomain disease targets in the search for effective new medical drugs. More ...
Contact:
Stefan Knapp, Buchmann Institut für Molekulare Lebenswissenschaften, Structural Genomics Consortium Frankfurt und Institut für Pharmazeutische Chemie, Campus Riedberg, Goethe University, Frankfurt/Main, Germany, knapp@pharmchem.uni-frankfurt.de
Publication:
Qin Wu, David Heidenreich, Stanley Zhou, Suzanne Ackloo, Andreas Krämer, Kiran Nakka, Evelyne Lima-Fernandes, Genevieve Deblois, Shili Duan, Ravi N. Vellanki, Fengling Li, Masoud Vedadi, Jeffrey Dilworth, Mathieu Lupien, Paul E. Brennan, Cheryl H. Arrowsmith, Susanne Müller, Oleg Fedorov, Panagis Filippakopoulos & Stefan Knapp (2019) A chemical toolbox for the study of bromodomains and epigenetic signaling. Nature Communications 10: 1915. http://dx.doi.org/10.1038/s41467-019-09672-2

