News Archive
Trapping light-catching proteins
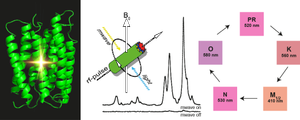
December 2017. The discovery of many new rhodopsins has shown that the typical structural scaffold provides the basis for a great variety of functions, including cation and anion pumps, sensors and channels. These also manifest themselves in different photocycle kinetics, which indicate important differences in their energy landscapes. These fine details need to be understood in order to comprehend how apparently similar molecular designs can yield such a variety of functions. The Glaubitz lab has therefore established a novel experimental setup in which photointermediate states can be cryo-trapped and analyzed by solid-state NMR. The sensitivity is enhanced by orders of magnitude though dynamic nuclear polarization. Photointermediates are created by sample illumination protocols. In the current study, photointermediates of proteorhodopsin (PR), the most abundant retinal protein on earth, have been investigated for the first time.
The data reveal unexpected conformational changes within the chromophore. Furthermore, two different M-states have been observed reflecting the Schiff base reorientation after the de-protonation step. The study provides novel insight into the photocycle of PR and also demonstrates the power of DNP-enhanced solid-state NMR to bridge the gap between functional and structural data and models. Full paper. JACS Spotlight.
Contact:
Clemens Glaubitz, Institute of Biophysical Chemistry and BMRZ, Riedberg Campus, Goethe University, Frankfurt am Main, Germany, www.glaubitz-lab.de
Publication:
Mehler M, Eckert CE, Leeder AJ, Kaur J, Fischer T, Kubatova N, Brown LJ, Brown RCD, Becker-Baldus J, Wachtveitl J, Glaubitz C (2017) Chromophore distortions in photointermediates of proteorhodopsin visualized by dynamic nuclear polarization-enhanced solid-state NMR. J Am Chem Soc 139:16143-16153. http://dx.doi.org/10.1021/jacs.7b05061)
Cluster of Excellence Macromolecular Complexes, Frankfurt am Main, Germany