News Archive
Study suggests paradigm shift about the way microbial rhodopsins function
April 2019. Proteorhodopsins are found in marine bacteria in various ecosystems and belong to the most abundant photoreceptors. Numerous proteorhodopsin-like sequences with specific properties have been found in different seas of the world. There are green proteorhodopsins and blue proteorhodopsins, colour-tuned to the depth of seawater in which they occur. Proteorhodopsins function as light-dependent proton pumps and convert light into a transmembrane, electrochemical gradient as a source of energy for the cell.
Like many microbial rhodopsins, proteorhodopsins have seven transmembrane helices and form oligomers. Salt bridge contacts control the formation of predominantly pentamers (consisting of five proteorhodopsin molecules) and a small fraction of hexamers (consisting of six molecules). It was not known whether these pentameric and hexameric arrangements have a direct functional relevance for proton transport.
Scientists from the Institute of Biophysical Chemistry at Goethe University Frankfurt now provide evidence for functionally relevant cross-protomer interactions within a proteorhodopsin pentamer. By combining solid-state NMR spectroscopy with dynamic nuclear polarization and light-induced cryotrapping of photointermediates, they found that a triad of specific amino acids interacts across the oligomerization interface. This triad participates in the photocycle and potentially acts as a pH sensor. These findings suggest a paradigm shift regarding the way microbial rhodopsins function.
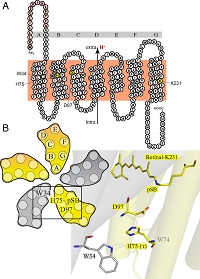
Figure legend: (A) Side view of a green proteorhodopsin molecule (protomer) showing its seven transmembrane helices (A-G) and the positions of the three amino acids W34 (tryptophan), H75 (histidine) and D97 (aspartic acid) that are involved in the triad. Protons are transported from the intracellular to the extracellular side. (B) Top view of a pentamer of five green proteorhodopsin protomers showing the positions of these three amino acids. Also shown are the detailed structural arrangements of the triad with the amino acid W34 in one protomer (gray) and the amino acids H75 and D97 in the neighboring protomer (yellow) (Proc Natl Acad Sci USA, DOI 10.1073/pnas.1817665116).
Contact:
Clemens Glaubitz, Institute of Biophysical Chemistry and Center for Biomolecular Magnetic Resonance, Riedberg Campus, Goethe University, Frankfurt/Main, Germany, glaubitz@chemie.uni-frankfurt.de
Publication:
Jakob Maciejko, Jagdeep Kaur, Johanna Becker-Baldus & Clemens Glaubitz (2019) Photocycle-dependent conformational changes in the proteorhodopsin cross-protomer Asp-His-Trp triad revealed by DNP-enhanced MAS-NMR. Proceedings of the National Academy of Sciences USA: published online on 4 April 2019. http://dx.doi.org/10.1073/pnas.1817665116
Cluster of Excellence Macromolecular Complexes